Cancer diagnosis using microarrays
DNA microarrays are a recent technology that has already had a huge impact on
biology and medicine. They are producing vast amounts of data which are
being analyzed to elucidate the inner workings of cells.
One of the most
powerful applications of this technique is in the diagnosis of cancer.
A common problem is that different types of cancer may look virtually
indistinguishable using conventional diagnostic techniques.
However the use of microarrays has been shown in many
cases to provide clear differential diagnosis rivaling or surpassing
other methods. At the moment this technique is too time consuming and
costly to be put into clinical practice. To achieve this goal it is useful
to drastically reduce the number of genes needed to accurately diagnose
a particular type of cancer.
Deutsch has developed a technique to analyze microarray data that shows how an accurate diagnosis
can be made by considering
far less genes than had been possible previously.
The software package "GESSES" can take microarray tumor data from many different types of
related cancer, and sift through it to produce a predictor based on a small
subset of genes. For example with data from small round blue cell tumors,
it was able to predict with 100% success rate test data using a predictor that
used less than 15 genes.
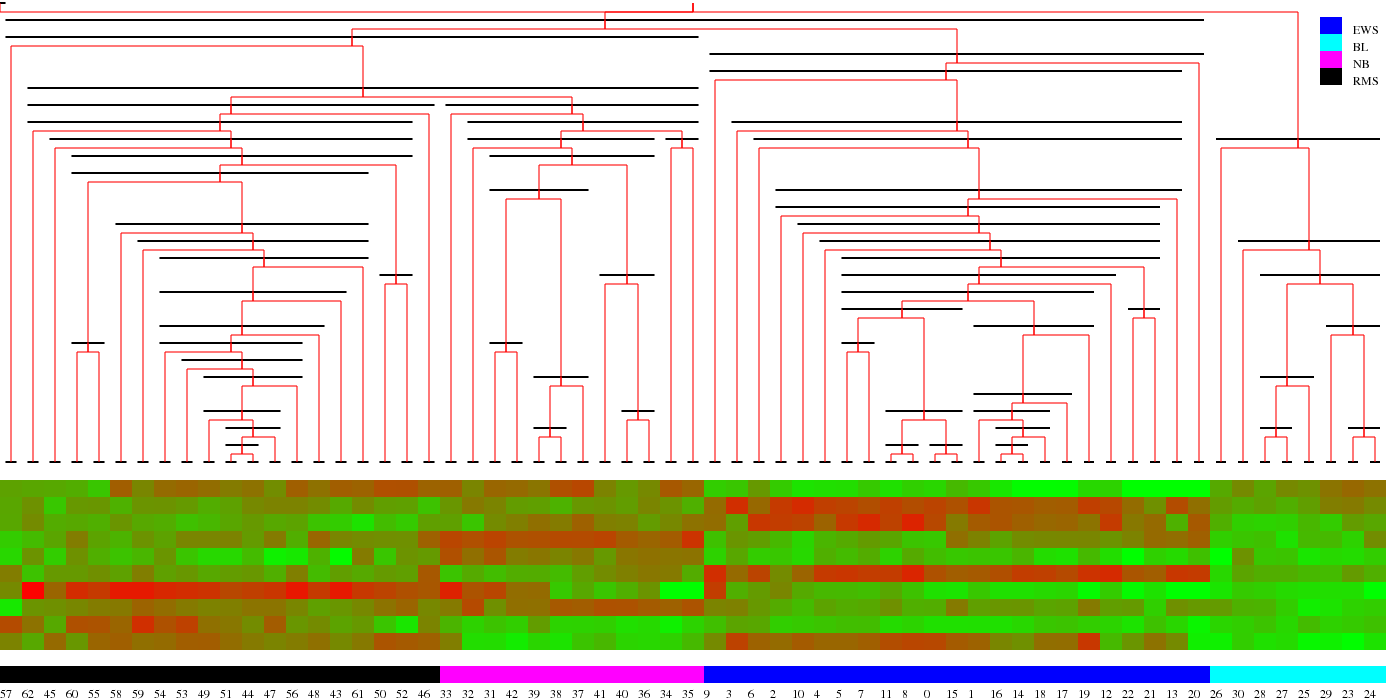
Above is the results of clustering the gene expression levels of patients using the genes picked out by GESSES. Improving microarrays using physicsMicroarrays have thousands of wells each of which contains many copies of a sequence of DNA. The mRNA of cells is amplified and chopped up and fluorescently tagged. Then this is put in contact with the microarray. The pieces of RNA that are exactly complementary to a well will bind to it and light up more if more of that kind of RNA is present. This is how it should work. But the physics of this are far more complicated and cause a substantial degradation in performance limiting the technique's resolution. In a nutshell the problem is this. If the temperature of the microarray is too high, nothing will bind. If it's too low, everything will. The microarray has to be close to the melting temperature to be useful. But the melting temperature for different strands of DNA or RNA depends on the sequence. To understand the binding, we have to actually model the physical binding of molecules in solution to molecules on the microarray. Coming up with such a model involves many effects and matching to experiment requires fitting because many crucial parameters have not been experimentally ascertained in this situation. Nevertheless, this can be done and gives a considerable improvement on the sensitivity of microarray data as compared to other techniques. |